Single cell RNA-Seq analysis in R tutorial is a powerful technique for studying the gene expression of individual cells. This tutorial will provide a comprehensive overview of the steps involved in single cell RNA-Seq analysis in R, from data preprocessing to data visualization.
Single cell RNA-Seq analysis has revolutionized the field of biology by allowing researchers to study the gene expression of individual cells. This has led to a number of important discoveries, including the identification of new cell types and the development of new treatments for diseases.
Single-Cell RNA-Seq Analysis in R Tutorial
Single-cell RNA-seq analysis is a powerful technique that allows researchers to study the gene expression of individual cells. This technique has revolutionized the field of biology and has led to new insights into the development, function, and disease of cells.
In this tutorial, we will provide a step-by-step guide to performing single-cell RNA-seq analysis in R. We will cover the following topics:
- Introduction to single-cell RNA-seq analysis
- Data preprocessing
- Data analysis
- Data visualization
- Case studies
Introduction to Single-Cell RNA-Seq Analysis: Single Cell Rna-seq Analysis In R Tutorial
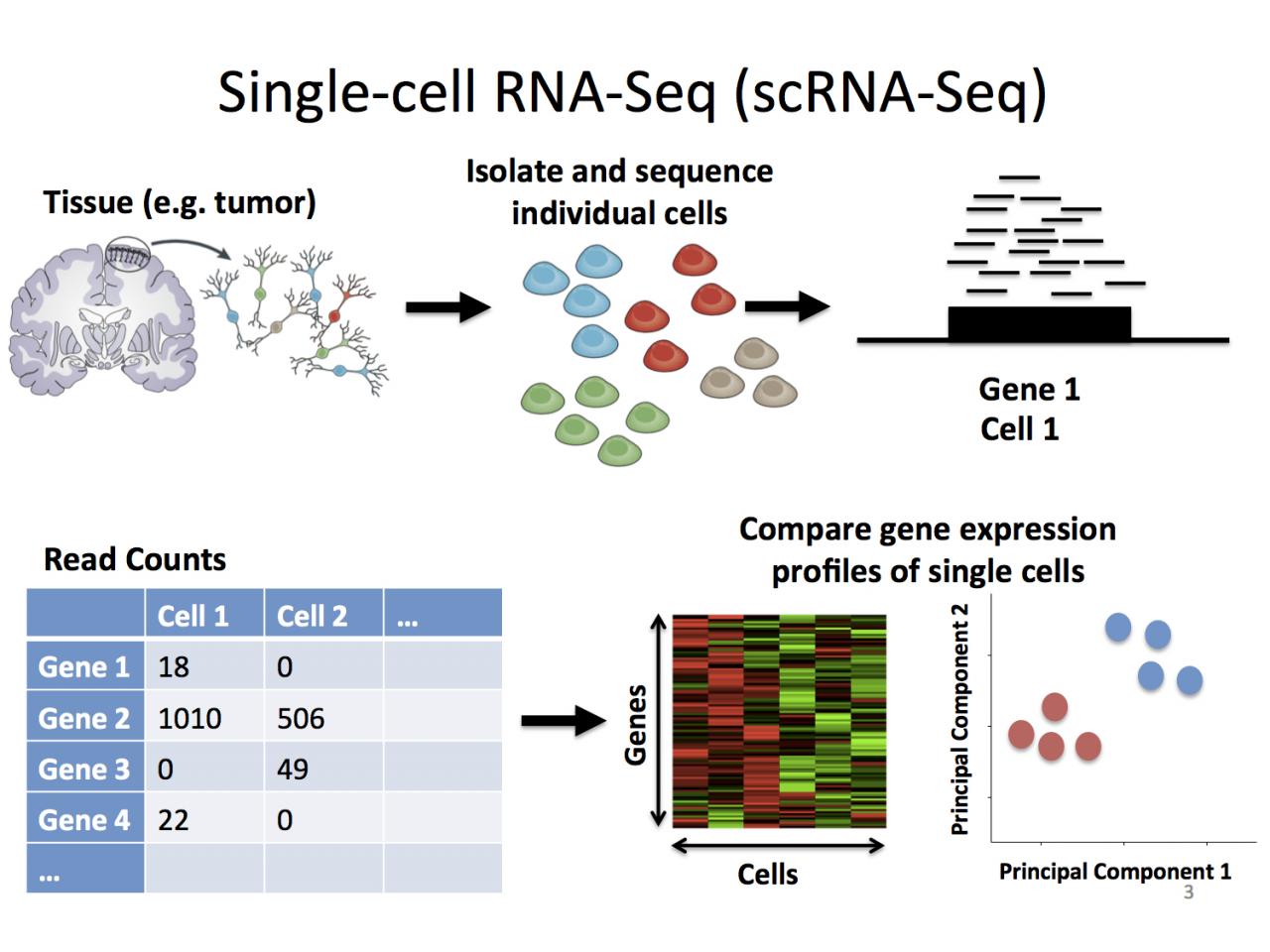
Single-cell RNA-seq analysis is a technique that allows researchers to study the gene expression of individual cells. This technique is performed by sequencing the RNA of individual cells and then analyzing the data to identify the genes that are expressed in each cell.
Single-cell RNA-seq analysis has a number of advantages over traditional bulk RNA-seq analysis. First, single-cell RNA-seq analysis allows researchers to identify cell-to-cell variability in gene expression. This information can be used to identify rare cell types and to study the development and differentiation of cells.
Second, single-cell RNA-seq analysis allows researchers to study the spatial organization of gene expression. This information can be used to identify the location of different cell types within a tissue and to study the interactions between cells.
However, single-cell RNA-seq analysis also has some limitations. First, single-cell RNA-seq analysis is a relatively expensive and time-consuming technique. Second, single-cell RNA-seq analysis can be difficult to interpret. The data is often complex and noisy, and it can be difficult to identify the genes that are truly differentially expressed.
Data Preprocessing, Single cell rna-seq analysis in r tutorial
The first step in single-cell RNA-seq analysis is to preprocess the data. This involves removing low-quality reads, aligning the reads to the reference genome, and counting the number of reads that map to each gene.
There are a number of different software packages that can be used to preprocess single-cell RNA-seq data. Some of the most popular packages include Cell Ranger, Seurat, and Scanpy.
The following code shows how to preprocess single-cell RNA-seq data using the Cell Ranger software package:
“`cellranger count
- -id=my_sample
- -transcriptome=refdata-cellranger-GRCh38-2020-A
- -fastqs=fastq_dir
“`
Data Analysis
Once the data has been preprocessed, it can be analyzed to identify the genes that are differentially expressed between different cell types.
There are a number of different statistical methods that can be used to identify differentially expressed genes. Some of the most popular methods include the t-test, the Wilcoxon rank-sum test, and the DESeq2 method.
The following code shows how to identify differentially expressed genes using the DESeq2 method:
“`dds <- DESeqDataSetFromMatrix(countData, colData(countData), design = design) dds <- DESeq(dds) results <- results(dds) ```
Data Visualization
Once the differentially expressed genes have been identified, they can be visualized using a variety of different methods.
Some of the most popular methods for visualizing single-cell RNA-seq data include heatmaps, scatterplots, and t-SNE plots.
The following code shows how to create a heatmap of the differentially expressed genes:
“`heatmap(results, show_row_names = FALSE, show_col_names = FALSE)“`
Closing Summary

This tutorial has provided a comprehensive overview of the steps involved in single cell RNA-Seq analysis in R. By following the steps Artikeld in this tutorial, researchers can use single cell RNA-Seq to study the gene expression of individual cells and make new discoveries about the biology of cells.


